LIGPLOT v.4.4.2
Operating manual
A program to plot schematic diagrams of protein-ligand interactions
Andrew C Wallace & Roman A Laskowski.
 Contents
Contents


 a. The algorithm
a. The algorithm b. Hydrogen bonds
b. Hydrogen bonds

 a. Running LIGPLOT
a. Running LIGPLOT b. Running LIGONLY
b. Running LIGONLY c. Showing atom accessibilities
c. Showing atom accessibilities







 a. Editing the PostScript file
a. Editing the PostScript file b. Editing ligplot.pdb using graphics
b. Editing ligplot.pdb using graphics c. Altering minimization parameters
c. Altering minimization parameters d. Same protein, different ligands
d. Same protein, different ligands



 a dimer or domain-domain interface
a dimer or domain-domain interface a. DIMPLOT for dimers
a. DIMPLOT for dimers b. Domain-domain interface
b. Domain-domain interface c. Defining protein domains
c. Defining protein domains d. Running DIMONLY
d. Running DIMONLY


![]() Appendix A - Brookhaven file format
Appendix A - Brookhaven file format


![]() Appendix B - LIGPLOT file formats
Appendix B - LIGPLOT file formats
 a. H-bonds file, .hhb
a. H-bonds file, .hhb
 b. Non-bonded contacts, .nnb
b. Non-bonded contacts, .nnb
 c. Residue CofM file, .rcm
c. Residue CofM file, .rcm


![]() Appendix C - The HBADD program
Appendix C - The HBADD program
 a. Pre-processor for HBPLUS
a. Pre-processor for HBPLUS
 b. The algorithm
b. The algorithm
 c. Problems
c. Problems


![]() Sample plots
Sample plots
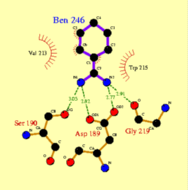
 a. Sample LIGPLOT outputs
a. Sample LIGPLOT outputs
 b. Sample DIMPLOT outputs
b. Sample DIMPLOT outputs
Last changed 7 Mar 2003.